Methyl Lysine Antibody, Agarose
|
Catalog#
Product Description
|
ICP0588
The methylated lysine antibodies are developed with methylated KLH and affinity purified with methylated lysine on agarose. The purified antibodies are covalently immobilized to beaded agarose. The product could be utilized as an affinity matrix for rapid isolation, purification, and enrichment of the proteins or peptide with methylted lysine residues, particularly trimethyl- and/or dimethyl-lysine residues.
|
|
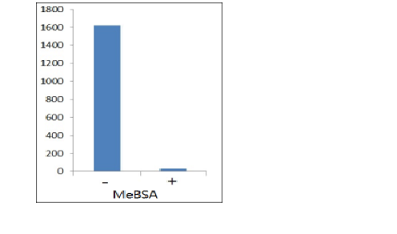
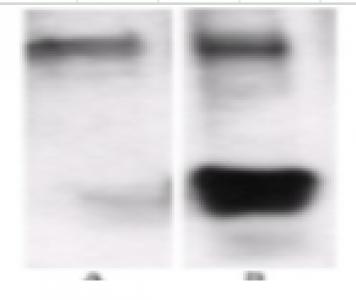
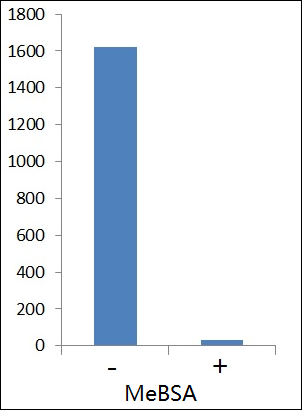
The ELISA titre (1:1620) of the methylated proteins eluted from 50 ul of the methyllysine-specific antibody affinity matrix (ICP0588) as detected with anti-methyllysine HRP (ICP0502). Approximately 50 mg of the swine spleen cell lysate were applied to the enrichment. ELISA signal can be inhibited methylated BSA (MeBSA).
|
|
Formulation
|
0.5 ml beaded agarose suspended in 1 ml slurry with glycerol
|
|
Antibody Immobilized
|
10 mg/ml of the methylated lysine antibodies are covalently linked to the agarose beads
|
|
Specificity
|
This antibody matrix selectively captures the peptides and proteins with methylated lysine residues (N-epsilon).
|
|
Binding Capacity
|
Approximately 0.5 mg of methylated histone/ml
|
|
Applications
|
Rapid isolation, purification, and affinity enrichment of peptides or proteins with the methylated lysine residues from mixtures of cell lysates or protease-digested samples.
|
|
Scientific Description
|
Protein methylation is a form of post-translational modification known to regulate many diverse biological processes. Detection, isolation and identification of the methylated protein/peptide is essential in proteomic studies. Affinity chromatography is one of the most efficient and rapid methods to enrich and purify the methylated species for further MS/MS identification.
|
|
Storage & Stability
|
Product is stable for 30 days at room temperature. For extended storage, store product at -20°C. Do not aliquot and shake thoroughly before use. Expiration date is one year from date of shipping of properly stored.
|
|
Product Specific References
|
-
1.Mol. BioSyst. 2013. 9: 2231-2247. doi: 10.1039/C3MB00009E
-
2.Biochem & Biophys Res. Com. 2014. 451 (2): 229-234. doi: 10.1016/j.bbrc.2014.07.11
-
3.Proteomics. 2015. 15 (13): 2166–2176. doi: 10.1002/pmic.201400521
|