|
Catalog#
Product Description
|
ICP0380
This pan-specific antibody is affinity purified using acetyl-lysine affinity chromatography. It recognizes proteins with acetylated lysine residues. The product has been utilized for proteomic studies of acetylated proteins and immunoaffinity chromatography separation and isolation of acetylated proteins and peptides from protease-digesting proteins of whole cells.
|
|
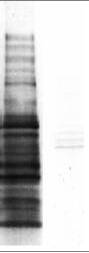
A B
Western blot analysis of the acetylated protein profile in HeLa cell lysate with anti-acetyl lysine antibodies (ICP0380, lane A) and with additional acetylated BSA (10 ug/mL) in the primary antibody (lane B).
|
|
Species
|
Rabbit
|
|
Formulation
|
250 μg/mL antibody in PBS, 50% glycerol
|
|
Immunogen
|
Acetylated KLH conjugates
|
|
Purification
|
The antibody was purified on acetyl-lysine agarose.
|
|
Specificity
|
The antibody recognizes proteins acetylated on lysine residues. Tested: acetylated histone, acetylated BSA, and acetylated MBP. There are no cross-reaction to non-acetylated proteins.
|
|
Applications
|
ELISA; WB (1:1000); IP; Immunofluorescence; Immunochemistry
|
|
Scientific Description
|
DNA transcription cannot take place unless DNA is unwound from the nucleosomes. The cell unwinds the DNA by acetylating lysine residues within the proteins. It has been suggested that acetylation of non-histone proteins (e.g., transcription factors) and histones may be involved. Acetylation of these proteins may result in signal transduction within the chromatic domains.
|
|
Storage & Stability
|
Product is stable for several weeks at 4°C. For extended storage, store product at –20oC. Expiration date is one year from date of shipping if properly stored.
|
|
Product Specific References
|
-
1. Mol. Cell. Proteomics. 2009. 8 (2): 215-225. doi:10.1074/mcp.M800187-MCP200.
-
2. Eur. J. Cell Biol.2011. 90 (2-3): 128-135. doi:10.1016/j.ejcb.2010.09.004.
-
3. Nucleic Acids Res. 2011. 39 (14): 5907-5925. doi:10.1093/nar/gkr162.
-
4. J. Lipid Res.2012. 53 (9): 1864 -1876. doi:10.1194/jlr.M026567.
-
5. J. Biol. Chem. 2012. 287 (39): 32307-32311. doi:10.1074/jbc.C112.403048.
-
6. Mol. Cell. Proteomics. 2012. 11 (5): 202-214. doi:10.1074/mcp.M112.017707.
-
7. Mol. Cell. Biol.2012. 32 (14): 2823-2836. doi:10.1128/MCB.00496-12.
-
8. PLoS Genet.2012. 8 (9): e1002948. doi:10.1371/journal.pgen.1002948.
-
9. Genes Dev.2012. 26 (13): 1473-1485. doi:10.1101/gad.193615.112.
-
10. Nature. 2012. 482 (7384): 251-255. doi:10.1038/nature10804.
-
11. Nature Med.2012. 18 (1): 159-165. doi:10.1038/nm.2559.
-
12. Mol. Syst. Biol.2012. 8 (1): 571. doi:10.1038/msb.2012.4.
-
13. J. Biol. Chem. 2012. 287 (29): 24460-24472. doi:10.1074/jbc.M112.382226.
-
14. J. Proteomics.2013. 79: 60-71. doi:10.1016/j.jprot.2012.12.001.
-
15. J. Proteome Res. 2013. 12 (9): 3952-3968. doi:10.1021/pr400245k.
-
16. Mol. Cell. Biol, 2013.33 (6): 1114-1123. doi:10.1128/MCB.01044-12.
-
17. Mol. Cell. Biol. 2013. 33 (19): 3864-3878. doi:10.1128/MCB.01495-12.
-
18. Mol. Microbiol. 2013. 89 (4): 660-675. doi:10.1111/mmi.12303.
-
19. PLoS ONE. 2013. 8 (7): e67513. doi:10.1371/journal.pone.0067513.
-
20. Proteomics. 2013. 13 (15): 2278-2282. doi:10.1002/pmic.201200072.
-
21. J. Biol. Chem. 2013. 288 (22): 15537-15546. doi:10.1074/jbc.M112.430207.
-
22. J. Biol. Chem. 2013. 288 (39): 28116-28125. doi:10.1074/jbc.M113.495549.
-
23. Nature. 2013. 496 (7443): 110-113. doi:10.1038/nature12038.
-
24. Diabetes. 2013. 62(10): 3404–3417. doi:10.2337/db12-1650.
-
25. PLoS Genet. 2014. 10(7): e1004490. doi:10.1371/journal.pgen.1004490.
-
26. Lipids. 2014. 49 (2): 119-131. doi:10.1007/s11745-013-3843-x.
-
27. PLoS ONE. 2014. 9 (4): e94816. doi:10.1371/journal.pone.0094816.
-
28. J. Biol. Chem. 2015. 290 (13): 8469-8481 doi:10.1074/jbc.M114.622696.
-
29. Mol. Cell. 2015. 59(5): 867-881. doi:10.1016/j.molcel.2015.05.006.
-
30. J. Biol. Chem. 2015. 290 (38): 23077-23093. doi:10.1074/jbc.M115.649806.
-
31. Plant Mitochondira: Methods and Protocols. 2015. 1305: 107-121. doi:10.1007/978-1-4939-2639-8_7.
-
32. Mol. Cell. Proteomics. 2015.14: 2429-2440. doi:10.1074/mcp.O114.047555.
-
33. Cell Death Differ. 2016. 23 (2): 279–290. doi:10.1038/cdd.2015.96.
-
34. Cancer Res. 2016. 76(13): 3802-3812. doi:10.1158/0008-5472.CAN-15-2498.
-
35. Methods Mol Biol. 2016. 1436: 85-94. doi:10.1007/978-1-4939-3667-0_6.
-
36. mSystems. 2016. 1(3): e00005-16. doi:10.1128/mSystems.00005-16.
-
37. J Biol Chem. 2016. 291(10): 5270-5277. doi:10.1074/jbc.M115.709428.
-
38. PNAS. 2016. 113(16): 4320-4325. doi:10.1073/pnas.1519858113.
-
39. Sci Rep. 2016. 6: 19722. doi:10.1038/srep19722.
-
40. PLoS ONE. 2016. 11(9): 1-16. doi: 10.1371/journal.pone.0162528.
-
41. The Journal of Biological Chemistry. 2016. 291: 5270-5277 doi: 10.1074/jbc.M115.709428.
-
42. Molecular & Cellular Proteomics. 2016. doi: 10.1074/mcp.O116.065219.
-
43. Histone Deacetylases. 2016. 1436: 85-94. doi: 10.1007/978-1-4939-3667-0 6.
-
44. Cancer Research. 2016. 76 (13); 3802-12 doi:10.1158/0008-5472.CAN-15-2498.
-
45. PLOS One. 2016. 11 (12): 1-20. doi: 10.1371/journal.pone.0168467.
-
46. American Society for Microbiology. 2016. 1 (3): 1-19. doi: 10.1128/mSystems.00005-16.
-
47. Scientific Reports. 2016. 6:31086: 1-9. doi: 10.1038/srep31086.
-
48. Scientific Reports. 2016. 6: 36013: 1-14. doi: 10.1038/srep36013.
-
49. South Dakota State University. 2016. 1-131.
-
50. CellPress. 2016. 167 (4): 985-1000. doi: http://dx.doi.org/10.1016/j.cell.2016.10.016.
-
51. Cell Death and Differentiation. 2015. 23: 279-290. doi: 10.1038/cdd.2015.96.
-
52. Oncotarget. 2016. 1-14. doi: 10.18632/oncotarget.12015.
-
53. Journal of Biological Chemistry. 2016. 1-24. doi: 10.1074/jbc.M116.744532.
-
54. Journal of Proteomics. 2016. doi: 10.1016/j.jprot.2016.12.006.
-
55. Molecular Microbiology. 2016. doi:10.1111/mmi.13595.
-
56. App. Environ. Microbiol. 2016. 83 (21) 1183-1195 doi: 10.1128/AEM.03056-15.
-
57. Journal of Thoracic Disease. 2016. 8 (9) 2485-2494. doi: 10.21037/jtd.2016.08.08.
-
58. EMBO Report. 2016. 17 (3) 455-469. doi: 10.15252/embr.201541132
-
59. Mol Cell Biochem. 2017. 432 (7) 7-24.doi: 10.1007/211010-017-2993-1
-
60. J. Proteom. 2017. 155: 63-72.doi: 10.1016/jprot.2016.12.006
-
61. Curr. Protoc. Protein Sci. 2017. 87: 14.11.1-14.11.18.doi: 10.1002/cpps.26
-
62. Biochimica et Biophysica Acta - Proteins and Proteomics. 2018. 1866 (3): 451-463. doi:10.1016/j.bbapap.2017.12.001
-
63. Nature Communications. 2018. 1039 (9). doi: 10.1038/s41467-018-03422-6
-
64. Front. Pharmacol. 2018. 201 (9). doi: 10.3389/fphar.2018.00201
-
65. Biochimica et Biophysica Acta - Porteins and Proteomics. 2018. 1866 (3): 451-463. doi: 10.1016/j.bbapap.2017.12.001
-
66. Front. Pharmocal. 2018. 201 (9). doi: 10.3389/fphar.2018.002
-
67. Nature. 2018. 1039 (9). doi: 10.1038/s41467-018-03422-6
-
68. Journal of Molecular Medicine. 2018.096 (3-4): 281-299. doi: 10.1007/s00109-017-1616-3
-
69. Molecular Microbiology. 2018. doi: 10.1111/mmi.13979
-
70. Journal of Biological Chemistry. 2019. 294(16): 6227-6239. doi: 10.1074/jbc.RA118.006051
-
71. Nature Communications. 2019. 10. doi: 10.1038/s41467-019-09024-0
-
72. Cancer Cell. 2019. 35(6): 916-931. doi: 10.1016/j.ccell.2019.05.002
-
73. American Journal of Physiology. 2019. 317(2). doi: 10.1152/ajpendo.00326.2018
|